About this tutorial

This tutorial provides an in depth lesson about mitochondrial DNA (mtDNA) and how it allows us to trace our maternal ancestry. The lessons start off with the basics, such as “what is mtDNA” and then advances to in depth case studies involving mtDNA. In order to fully understand the power of mtDNA testing and its applications in ancestry, it is beneficial to understand the science behind the technology and have a good idea of how ancestry testing works, including understanding its strengths and limitations.
This tutorial consists of a number of lessons that will dissect the mtDNA, allowing you to learn the details of mtDNA markers and hopefully give you a full technical understanding of how mtDNA ancestral tests work.
You don’t need to understand how mtDNA testing works in order to understand your maternal ancestry test results, but the more you know about “how”, “why” and “what’s next” when it comes to mtDNA testing, the more you will get out of your mtDNA ancestry testing experience.
What is mtDNA?

mtDNA stands for “mitochondrial DNA”. All of us, both males and females, carry mtDNA. mtDNA is found in most of the cells in our body.
mtDNA is unique because while most of the DNA in our body is found in the nucleus of our cells, mtDNA is found in small structures or organelles called “mitochondria”. Mitochondria are found in the cytoplasm of our cells, NOT in the nucleus (remember this, because it’s important when we discuss how mtDNA is inherited).
Mitochondria is important for producing energy “ATP” for our cells.
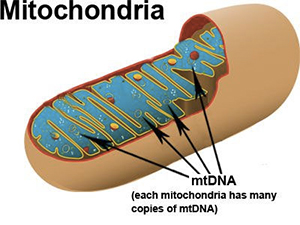
Many copies of mtDNA are found in every mitochondria and many mitochondria are present in the cytoplasm of each cell. That means that we have many more copies of mtDNA in our cells than other types of DNA which are present in only one set per cell. The huge abundance of mtDNA as well as its small size makes it an excellent candidate for forensic studies of old or degraded samples. Many archaeological studies of ancient DNA samples which are hundreds of years old rely on mtDNA testing.
mtDNA has a unique inheritance pattern.
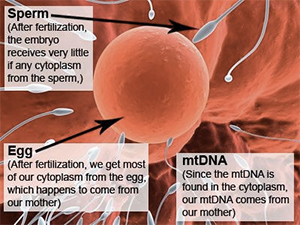
mtDNA has a very unique inheritance pattern which differs from all the other types of DNA in our body. We inherit all of our mtDNA from our mother, and our mother inherited all of her mtDNA from her mother, and so on. It is passed down strictly along the direct maternal line from a mother to all of her children. Males will carry the mtDNA of their mother, but when they have children, their children will carry the mtDNA of their own mother, not their father. Thus, only daughters will pass the mtDNA on to future generations.
The reason for the maternal inheritance pattern of mtDNA is due to its localization in the cytoplasm of the cell. When an egg is fertilized, the cells of the resulting embryo contain the cytoplasm of the egg, not the sperm. Since mtDNA is found only in the cytoplasm, all of our mtDNA comes from our mother, not our father. As the embryo continues to develop into a full grown human, all of the cells in the resulting human contain strictly the cytoplasm and mtDNA of the mother, not the father.
Why does mtDNA hold maternal ancestral information?
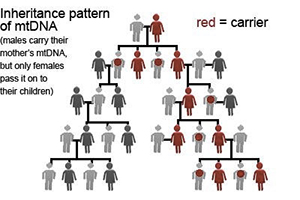
The maternal inheritance pattern of the mtDNA has important significance for ancestral studies. While most of the other types of DNA in our body become mixed as they are passed down from generation to generation, mtDNA remains unmixed because it follows a strict single line of descent from mother to child. This means that our mtDNA is the same as our mother’s and our mother’s mother’s mtDNA from hundreds, even thousands of generations ago.
By testing our own mtDNA, we are able to indirectly read the mtDNA genetic code of our own maternal ancestors from thousands of generations ago. mtDNA testing will allow us to trace our direct maternal lineage (mother’s mother’s mother’s….. maternal lineage).
Facts about mtDNA
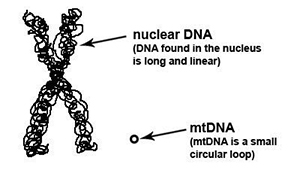
A good understanding of the basics of mtDNA will help you to better understand mtDNA ancestry discussions in this tutorial.
What does mtDNA look like?
1. It’s round! Unlike the other DNA types in our body which are linear, mtDNA happens to be a round circle, called a “plasmid”.
2. It’s small! While nuclear DNA (DNA found in the nucleus of the cell) is a staggering 49,530,000 to 247,200,000 bases pairs in length, mtDNA is only approximately 16,569 to 16,571 base pairs in length. Don’t worry if you don’t know what a “base pair” is – we will be talking about base pairs in more detail later in this lesson.
Why is mtDNA so different from all of the other DNA in our body?

The strange appearance of mtDNA in comparison to the other DNA types in our body has something to do with its ancient origins. Mitochondria has many of the same features as single cell organisms called “prokaryotes”. Bacterial cells are prokaryotes. The mtDNA that is found inside the mitochondria is a circular plasmid, just like the DNA in bacteria.
The “endosymbiotic hypothesis” suggests that the reason for the extremely close resemblance of mtDNA to bacterial DNA is that 1.7 to 2 billion years ago, mitochondria were originally bacteria that were “engulfed” by a cell and became permanently incorporated in the cytoplasm of the cell. This is called a “symbiotic” relationship because the cell and the bacteria provided a survival advantage to each other (mitochondria produces energy “ATP” for the cell, and the cell provides protection). This explains why the mtDNA is small and circular and found in the cytoplasm instead of the nucleus of the cell.
What does mtDNA do? What is its function?
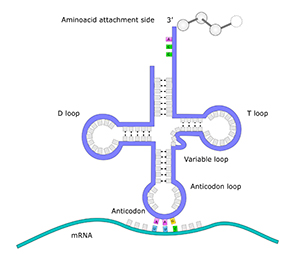
mtDNA contains the genetic code for at least 37 essential genes. 13 of the genes are responsible for producing proteins, 22 of the genes hold the genetic code to produce transfer RNA (tRNA), and two genes encode ribosomal RNA (rRNA). Thus, the mtDNA is very important, and when something goes wrong with the mtDNA, it can lead to mtDNA diseases such as exercise intolerance, Kearns-Syre syndrome and even death.
The size, structure and importance of mtDNA for survival, all play a role in where the majority of ancestral markers are located in the mtDNA and will allow you to understand the testing methods used to detect ancestral markers in the mtDNA.
mtDNA Structure
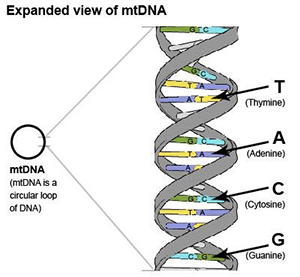
Understanding the structure of mtDNA will help you to understand the different types of mtDNA tests available.
mtDNA is a circular loop of DNA. DNA is the chemical that carries genetic information. DNA looks like a long ladder twisted into a “double helix”. The sides of the ladder are the ”backbone”, and the rungs of the ladder consist of “nucleotide bases”. There are 4 types of bases: A, C, T, and G. “A” is always connected to “T”, and “C” is always connected to “G”. The pairing of A from one strand with T from the other strand and the pairing of C from one strand with the G from the other strands leads to the term “base pairs”.
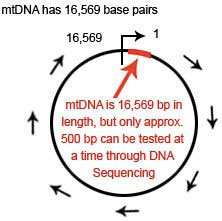
mtDNA is a circular loop of DNA which is 16,569 base pairs in length
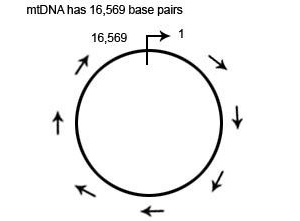
The mtDNA loop is 16,569 base pairs in length. The location of each base pair in the mtDNA can be specified with an accession number according to its position in the mtDNA. When numbering the base pairs, we start at the “origin”. The origin is arbitrarily located in the D-Loop between the HVR1 and HVR2 regions.
The position of any base pair in the mtDNA is relative to the origin. The position of any base pair in the mtDNA is designated by counting from “1″ clockwise around the mtDNA. Thus, the positions are named 1 to 16,569 (remember this because it is important when we start talking about ancestral markers).
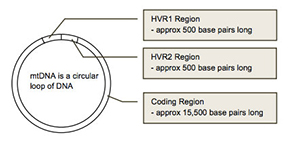
mtDNA contains three different regions
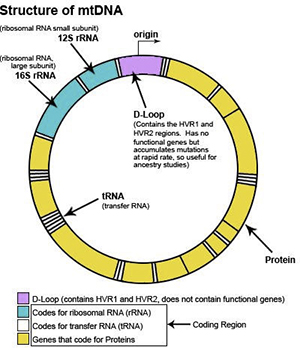
mtDNA can be divided into three main regions: HVR1, HVR2 and Coding Region. The HVR1 region is approximately 500 nucleotides in length (spans location 16,000 to 16,569). The HVR2 region is approximately 400 nucleotides in length (spans locations 1 to 400). The coding region is approximately 15,500 nucleotides in length (spans location 400 to 16,000).
The HVR1 and HVR2 regions of the mtDNA contain the highest concentration of ancestral markers and are the most common starting point for maternal ancestral studies. The HVR1 and HVR2 regions are considered non-vital parts of the mtDNA because they do not have a useful biological function. Thus, whenever a mutation occurs in the HVR1 or HVR2 region of one of our ancestors (we will discuss ancestral markers and mutations later), the individual does not die and survives to pass the mutation along to all future generations.

In contrast, the coding region contains many important genes which are essential for the survival of the individual, so whenever a mutation occurs in the coding region, it is often lethal and the organism dies. Thus, very few mutations which occur in the coding region can be passed down to future generations. For this reason, over a period of tens of thousands of years, many mutations have accumulated in the HVR1 and HVR2 regions, but a much smaller number of mutations can be found in the coding region.
When tracing ancestry, scientists usually begin by testing the HVR1 and HVR2 regions because of its small size and abundance of mutations or “ancestral markers”. The coding region also contains extremely important ancestral markers but it is a much larger region and more expensive to test. Testing all three regions of the mtDNA is called “mtDNA Full Sequencing” and provides the most comprehensive analysis of an individual’s maternal ancestry.
What is an ancestral marker?
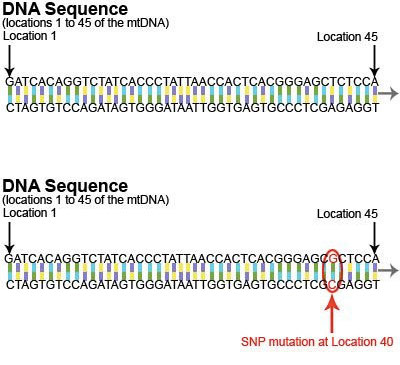
mtDNA is a circular genome consisting of 16,569 pairs of nucleotides. The diagram here illustrates a short section (bases 1-45) of the mtDNA genome. As shown in the diagram, the mtDNA genome exists as double-stranded DNA. This image shows the mtDNA reference sequence (top) and a variant mtDNA sequence (bottom).
Ancestral markers are “mutations”, little changes or “hiccups” that naturally occur in the genetic code of the mtDNA. There are many types of mutations, but the type of mutation most commonly found in mtDNA is called a “SNP” (single nucleotide polymorphism). A SNP mutation occurs when a single nucleotide is replaced with a different nucleotide. For example, in this diagram, the “T” at location 40 is replaced by a “G”.
This mutation is documented as follows:
Location: 40
Nucleotide Change: T>G (also indicated as T40G)
When you test your mtDNA, your results report will document the variations that you carry in your mtDNA.
Let’s take a look at a sample report:
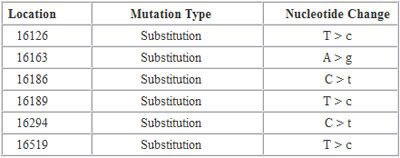
In this report, “Location” refers to the locations on the mtDNA where a mutation has been detected. “Mutation Type = Substitution” means that a nucleotide has been substituted by a different nucleotide. “Nucleotide Change” indicates what was substituted. Let’s take a look at the first mutation in the list. Location “16126″ means that a mutation has been detected at location “16126″. T>c means that a “T” has been replaced by a “C” at this location.
A second way to look at your results is to take a look at the actual sequence. In this example, the sequence shows the results for the HVR1 Test, encompassing locations 16001 to 16520 of the mtDNA. Remember, only one of the two strands in the pair is shown when reporting the sequence!
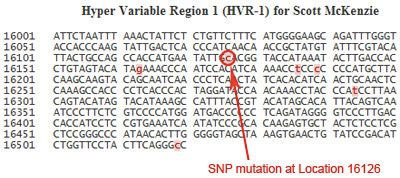
Nucleotides are listed one line at a time, from left to right. In this example, the first line shows the results for locations 16001 to 16050, the second line shows the results for 16051 to 16100, and so on. Mutations in the sequence are highlighted in pink. In this sequence, the nucleotide at location 16126 is highlighted in pink, indicating that a mutation is detected here, and the nucleotide has been replaced by a “C”.
The unique set of mutations that you carry in your mtDNA holds information about your maternal ancestry.
Detecting mutations in the mtDNA
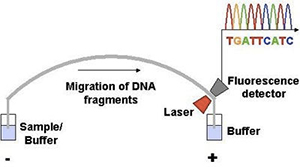
A basic understanding of DNA testing techniques will help you to understand the science behind DNA ancestry testing.
The entire genetic sequence of the mtDNA region tested is uncovered using a testing method called “Sanger Sequencing”. Sanger sequencing is a special process which is used to read the chain of nucleotides in a specific segment of your DNA, much like reading a book.
This technology allows the lab to read the entire genetic code of a whole section of your mtDNA. The following report is an example of the results of a sequencing test in the HVR1 region of an individual’s mtDNA.

As you can see, all of the nucleotides in HVR1 region (locations 16001 to 16520) have been decoded. All mutations detected in the sequence are indicated in pink.
The benefit of Sanger Sequencing technology is that it can accurately read entire lengths of your mtDNA and represents the most comprehesive way to test mtDNA. The drawback of Sanger Sequencing technology is that only approximately 400 to 800 nucleotides can be read at a time (in one run) so it is very expensive to perform.
Sanger Sequencing technology is the most comprehenstive way to detect mutations in the mtDNA. The HVR1 and HVR2 reigons are the most widely studied region of the mtDNA for ancestral studies and a great starting point for beginner genetic genealogists.
The coding region of the mtDNA is very expensive to sequence due to its large size. While the coding region does not have a high concentration of mutations, the mutations that it does carry are highly informative. Testing the mutations in the coding region is necessary in order to confirm an individual’s haplogroup and subclade. Once the HVR1, HVR2, and coding regions are fully sequenced, the entire mtDNA genome has been sequenced and no further mtDNA testing will ever be required.
Sequencing all three regions allows the laboratory to conclusively confirm your mtDNA haplogroup and mtDNA subclade. Furthermore, as more mtDNA data becomes available and the mtDNA tree continues to grow, you will continue to receive the latest classifications and updated results if all three regions of your mtDNA have been sequenced.
Tracing ancestry with mtDNA

Let’s take a look at how the mutations in our mtDNA act as ancestral markers, allowing us to trace our maternal ancestry.
We all have a unique pattern of SNP mutations in our mtDNA. Our SNP mutations can be used to trace our maternal ancestry in two ways – by direct comparisons and by mtDNA haplogroup and subclade determination.
1. Direct Comparisons:
By testing your mtDNA, you will discover the unique set of mutations that was passed down to you from your maternal ancestors along your direct maternal line. Your mtDNA “profile” is the unique set of mutations that you inherited from your own mother, and it is unique to your maternal ancestry. For example, all individuals living anywhere in the world today who have descended from the same maternal lineage as you will have exactly the same mtDNA profile as you (i.e. you are linked through a common maternal ancestor). If the mtDNA test shows that someone has a completely different mtDNA profile as you, that means that he/she definitely did not descend from the same maternal lineage as you (i.e. you are not directly linked on your direct maternal line). Once you test your mtDNA markers, you can:
- Compare your mtDNA profile to other individuals to determine whether you may have descended from the same maternal lineage. A mismatch will conclusively show that you definitely did not descend from the same maternal lineage. This test can confirm or refute family legends where the maternal lineage of different family branches are in question.
- Use your markers to search the DNA Reunion database to find other individuals from around the world who share the same mtDNA markers as yourself to find possible family links.
The more regions of your mtDNA that you compare, the more stringent your comparison will be. Your mtDNA contains three regions – the HVR1, HVR2 and coding region. Comparing all three regions of the mtDNA will provide a much more stringent match than comparing only the HVR1 or HVR2 regions. Please remember that in order to compare two individuals using all three regions, both individuals compared must have tested all three regions. If you have tested three regions and the person that you are comparing against had only tested one region, then you can only compare one region.
2. mtDNA Haplogroup Determination:
mtDNA studies have shown all people living today can be traced back to a common maternal ancestor who lived in Africa approximately 150,000 years ago. Over time, many different ancient family groups “haplogroups” eventually journeyed out of Africa and populated the rest of the world. mtDNA haplogroups (labeled A to Z) are associated with unique ancient migration routes out of Africa which led to different regions of the world (the paternal line equivalent is the “Y-DNA haplogroup).
The maternal ancestry test allows you to determine which mtDNA haplogroup you belong to. Once you find out which mtDNA haplogroup you belong to, you can find out which general region of the world your maternal ancestors came from. Please note that mtDNA haplogroups are NOT country specific. There are no haplogroups which are found in only one country and not a neighboring country.
mtDNA haplogroups can be further classified into finer sub-branches called “subclades”. Your mtDNA subclade can only be determined by sequencing all 3 regions of your mtDNA: HVR1, HVR2 and coding region. Knowing your subclade can often provide further geographical localization of your ancestry, if published research on the geographical distribution of the subclade is available.
The following chart shows the mt-DNA haplogroups found in each region.
| Region/Population | Major mt-DNA haplogroups |
| Native Americans | A, B, C, D, X |
| Oceanic and Aboriginal Australians | P, Q, R, S |
| East Asian | A, B, C, D, E, F, G, M, Y, Z |
| South Asian (i.e. India) | G, M, R, W |
| Europe and Middle East | H, HV, HV0, I, J, JT, K, R0, T, U, V, W, X |
| African | L0, L1, L2, L3, L4, L5, L6 |
mtDNA Test Types
In this section, we will provide a broad overview of the different types of mtDNA tests available and go over some case studies to help you understand when they are used and what each one will tell you about your ancestry.
Your mtDNA consists of three regions: HVR1, HVR2, and coding regions. Ancestral markers are found in all three regions of your mtDNA. mtDNA testing focuses on uncovering the markers in each of the three regions of the mtDNA:
There are three main types of mtDNA tests:
1. HVR1 Test (sequencing entire HVR1)
2. HVR1 and HVR2 Test (sequencing entire HVR1 and HVR2 region)
3. HVR1, HVR2 and Coding Region Test, aka “mtDNA full sequencing test” (sequencing the entire mtDNA, which includes the HVR1, HVR2 and coding region)
Let’s take a look at the main differences between these test types:
Test Type #1: HVR1 Test
Overview:
The HVR1 Test is the most fundamental mtDNA test, and it is always the first test that is performed when you start tracing your maternal ancestry. The HVR1 test uses DNA sequencing technology to read all of the nucleotides from locations 16,000 to 16,400 of your mtDNA. This is the entire HVR1 region, located in the D-Loop of the mtDNA.

Highlights of the HVR1 Test:
The HVR1 region is considered the most informative region of the mtDNA for ancestral studies for a number of reasons:
– The HVR1 region is located in the D-Loop, so it contains an extremely high concentration of mutations (ancestral markers). Thus, this region is highly variable and is good for checking for matches with other individuals.
– The entire HVR1 region can be easily tested using sequencing technology, as all 400 nucleotides in the entire HVR1 region can be read from a single test.
– The HVR1 region is the most well studied region of the mtDNA due to its high concentration of mutations and ease of testing. Most scientific studies to date, including indigenous data and other anthropological studies, have focused mainly on the HVR1 region. Thus, there is more scientific data available for markers in the HVR1 region than any other region of the mtDNA, making the HVR1 region by far the most informative region of the mtDNA.
Prerequisite:
There is no prerequisite for taking the HVR1 test. The HVR1 test is always the first and most fundamental test that is performed when using mtDNA to trace ancestry. The HVR1 test can be used “stand-alone” for search and comparisons. All of the other test types serve to supplement the results of the HVR1 test. If you are not a match to another individual in your HVR1 region, it can conclusively show that you are definitely not descendants from the same maternal lineage. If you are finding matches with another individual at the HVR1 region, you can consider upgrading to the HVR2 and coding region tests to increase the stringency of the comparison.
Test Type #2: HVR2 Test
Overview:
Like HVR1 testing, the HVR2 test also uses DNA sequencing technology. The HVR2 test focuses on reading all of the nucleotides from locations 1 to 400 of the mtDNA. This is the entire HVR2 region, the second most important region in the D-loop of the mtDNA.

Highlights of the HVR2 Test:
Like HVR1, the HVR2 region is also located in the D-Loop of the mtDNA so it contains many ancestral markers. The HVR2 region is always tested in conjunction with, or subsequent to HVR1 Testing. The HVR2 test will supplement the HVR1 results in the following ways:
– Strengthen the results of searches and comparisons, as the more regions of the mtDNA that are used for comparison, the more stringent and precise the results of the comparison.
– Prerequisite for the coding region test. The HVR2 region contains quite a few markers which are important for subclade determination, and when used in conjunction with HVR1 and coding region tests, will allow you to determine your mtDNA subclade.
Prerequisite:
The HVR1 test is a prerequisite for taking the HVR2 test. The HVR2 region is rich in ancestral markers and HVR2 testing is an excellent way to supplement the results of the HVR1 Test.
Test Type #3: mtDNA Coding Region Test
Overview:
The mtDNA Coding Region Test is a full sequencing test which sequences the entire coding region of the mtDNA. Sequencing the HVR1, HVR2 and coding region of the mtDNA will fully uncover the entire mtDNA sequence and no further mtDNA tests will ever be required. Sequencing the HVR1, HVR2, and coding region will allow you to conclusively confirm both your mtDNA haplogroup as well as your mtDNA subclade. Individuals who have fully sequenced all three regions of their mtDNA will receive constant upgrades at no charge whenever new SNPs are discovered and the mtDNA subclade becomes further refined.

Prerequisite:
The HVR1 and HVR2 tests are prerequisites for the Coding Region Test.
The Cambridge Reference Sequence
The Cambridge Reference Sequence (CRS) is a fundamental part of mtDNA data analysis. A basic understanding of the CRS and how it is used in determining mutations will allow you to understand how the mutations in your results report were derived.
What is the CRS?
The CRS is the first human mtDNA that was ever fully sequenced and published. The work was performed by scientists at Cambridge University, and this groundbreaking study was officially published in 1981. Click here to view a copy of the original publication.
This publication represents the first time that the mtDNA was sequenced. The donor whose DNA was used for this ground-breaking project was of European descent and belonged to European mtDNA haplogroup H, subclade H2a2a.
Since this was the first mtDNA sequence ever published, this sequence was thereafter referred to as a “reference sequence” upon which all further mtDNA sequences from labs around the world are compared to. This original sequence eventually came to be known as the “Cambridge Reference Sequence” and all mtDNA which is sequenced is compared to the CRS.
Mutations are determined based on comparison with CRS
When we state that we have variations or mutations in our mtDNA, we are actually identifying regions of our DNA which differ from the CRS. Let’s take a look at an actual mtDNA variation report:

In this report, the HVR1 region was tested, and six variations were detected, indicating that this individual’s HVR1 region differs from the CRS at six different locations. Let’s take a look at the first variation in the list: 16126 T>c. This means that the individual’s mtDNA is different from CRS at location 16126. It shows that CRS has a “T” at this location, but the person tested has a “C”.
Let’s look at the same results based on the sequencing report:

All of the letters in black are the same as CRS. All of the nucleotides in red are different from CRS and are considered “variants”.
The key point to remember is that when the results of mtDNA testing are used for genealogical purposes, the results are compared to the CRS and mutations are reported as “differences” between the results and the CRS. However, this can lead to confusion for beginner genetic genealogists because instinctively, people usually think that when scientists look for mutations, they should be comparing our mtDNA to that of the earliest human DNA to see how our DNA has changed over time. However, that is not how the research community had decided to approach the mtDNA. The consensus within the scientific community was that mtDNA would always be compared to the CRS. Since this is the case, it is important for you to become familiar with how this “reverse” method is used to analyze our mutations and determine haplogroups.
More recently, RSRS became a recognized method to classify variants in the mtDNA. The RSRS method uses the origin as the root and compares your results against the origin.
What are mtDNA haplogroups?
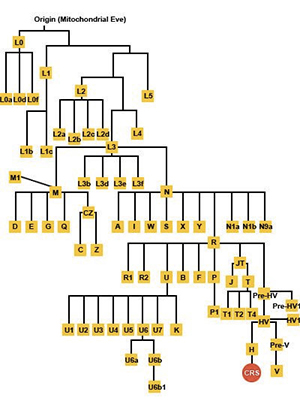
A phylogenetic tree generated using mtDNA SNPs shows how all people living today descended from a common ancestor (Mitochondrial Eve) who lived in Africa over 150,000 years ago. Every person living today can trace his/her ancestry to a branch of this tree, called a haplogroup. The European individual whose mtDNA sequence is famously called the CRS is located at a distant branch of the tree as shown in the diagram below:
Now, let’s take a look at how your mtDNA haplogroup is determined.
To determine your mtDNA haplogroup, always start with the CRS and move away.
Example #1: If you HAVE mutations at locations 263 and 7028, and DO NOT have mutations at locations 14766 or 16067 or 16298, then you belong to Haplogroup HV:

Example #2: If you HAVE mutations 263, 7028, 14766, 73, 11251, 16126, and 16069, and DO NOT have a mutation at 16294, then you belong to Haplogroup J.

Example #3: If you HAVE mutations 263, 7028, 14766, 73, 11719, 12705, 16223, 10873, 2352, and 150, then you belong to Haplogroup L3e:

Summary of procedure for determining mtDNA Haplogroups:
To determine your haplogroup, always start from the CRS and move backwards in the tree to see which mutations you have and which ones you do not have. Your haplogroup is determined by the difference between your markers versus CRS.
What if I don’t have any mutations?
If you do not have any mutations, that means that your mtDNA sequence (at least the part that was tested) is exactly the same as the CRS. The CRS belongs to a branch of haplogroup H, so if you belong to haplogroup H, chances are that you will not have many mutations in comparison with the CRS.
How are mtDNA haplogroups determined?

mtDNA haplogroups are determined by examining the pattern of SNP mutations in your mtDNA. SNP mutations are small “mistakes” that occur naturally in your DNA. SNP mutations are rare, occurring at a rate of approximately one mutation every few hundred generations. However, once a mutation occurs, it acts as a “time-and-date-stamp”, because it is passed on to all future generations. Each mutation event can be linked to a time and place in history, and by testing the mutations in your mtDNA, you can determine your mtDNA haplogroup.
Let’s take a look at how mutations can allow us to determine our haplogroup and retrace the ancient migration path of our ancestors using the following hypothetical example:
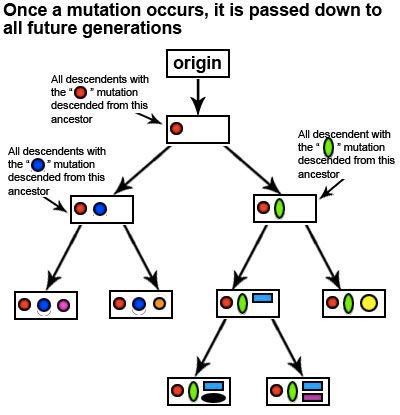
As you can see from this diagram, whenever a new marker occurs, it is passed down to all future generations. By studying the pattern of markers that appear in various indigenous populations from around the world, scientists have a general idea of where and when each marker first appeared. The pattern of markers have allowed scientists to generate a mtDNA phylogenetic tree. The main branches of the tree are called haplogroups and the finer sub-branches of the tree are called subclades. Each haplogroup and subclade can be associated with specific regions of the world.
What are mtDNA Subclades?
All people living today can trace their maternal ancestry back to one of 26 core mtDNA haplogroups. Haplogroups are the main branches of the mtDNA phylogenetic tree and represent extremely ancient family groups, which arose tens of thousands of years ago. Over time, the descendants of each haplogroup formed further subgroups, called subclades. Once you discover which mtDNA haplogroup you belong to, you can further fine tune your results by tracing which sub-branch (subclade) of your haplogroup you belong to through sequencing the coding region of your mtDNA.
Subclades are named using numbers and letters. For example, the subclades of haplogroup H include H1, H2, H3, H4,…. and so on. The subclade H2 can be further classified as H2a, H2b, etc. Similarly, the subclade H5 can be further classified as H5a, H5b, etc.
If you have tested all three regions of your mtDNA (HVR1, HVR2 and coding region), your mtDNA haplogroup and subclade will be confirmed. The mtDNA phylogenetic tree is constantly expanding as new subclades with greater resolution are being discovered at a rapid rate. Individuals who have sequenced all three regions of their mtDNA will automatically receive the latest subclade designation as the mtDNA tree grows over time.